A group of scientists in India is working on genomic sequences of SARS-CoV-2 around the World, including India, to identify genetic variability and potential molecular targets in virus and human to find the best possible answer to combat the COVID 19 virus.
Breaking down the novel coronavirus challenge into many pieces to get to its root and see it from multiple directions, Dr. IndrajitSaha, Assistant Professor in the Department of Computer Science and Engineering of National Institute of Technical Teachers’ Training and Research, Kolkata and his team have developed a web-based COVID- Predictor to predict the sequence of viruses online on the basis of machine learning and analysed 566 Indian SARS-CoV-2 genomes to find the genetic variability in terms of point mutation and Single Nucleotide Polymorphism (SNP).
The study being sponsored by Science and Engineering Research Board (SERB), a statutory body under the Department of Science and Technology (DST), has been published in the Journal called Infection, Genetics, and Evolution. They have mainly found that 57 out of 64 SNPs are present in 6 coding regions of Indian SARS-CoV-2 genomes, and all are nonsynonymous in nature.
They have extended this research for more than 10 thousand sequences around the globe, including India and found 20260, 18997, and 3514 unique mutation points globally, including India, excluding India and only for India, respectively.
The scientists are on the track to identify the genetic variability in SARS-CoV-2 genomes around the globe including India, find the number of virus strains using Single Nucleotide Polymorphism (SNP), spot the potential target proteins of the virus and human host based on Protein-Protein Interactions. They also carried out integrating the knowledge of genetic variability, recognise candidates of synthetic vaccine based on conserved genomic regions that is highly immunogenic and antigenic and detect the virus miRNAs that are also involved in regulating human mRNA.
They have computed the mutation similarity in sequences of different countries. The results show that the USA, England, and India are the top three countries having the geometric mean, 3.27%, 3.59%, and 5.39%, respectively, of mutation similarity score with other 72 countries. The scientists have also developed a web application for searching the mutation points in SARS-CoV-2 genomes globally and country wise. Besides, they are now working more towards protein-protein interactions, epitopes discovery, and virus miRNA prediction.
[The link of the COVID-Predictor
http://www.nitttrkol.ac.in/indrajit/projects/COVID-Predictor/index.php
Link to publicationhttps://doi.org/10.1016/j.meegid.2020.104457.
The link to the work on mutations which is under review now.
Link: http://www.nitttrkol.ac.in/indrajit/projects/COVID-Mutation-10K/
For more details, please contact: Dr. IndrajitSaha, NITTTR, Kolkata,
Email: indrajit@nitttrkol.ac.in, Mob: 9748740602]
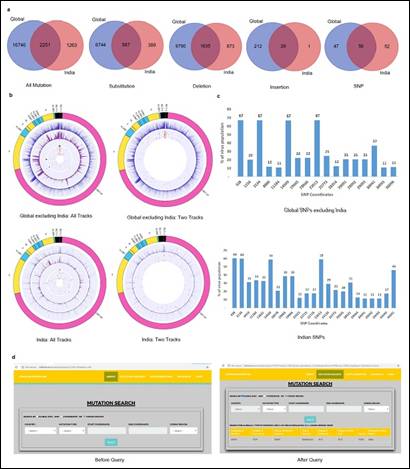
- Venn Diagrams between Global without India and India for all unique Mutation, Substitutions, Deletions, Insertions and SNP, (b) BioCircos plots to illustrate the frequency of Mutations across the Global excluding India and Indian SARS-CoV-2 genomes through different tracks, e.g., Substitution as outer track 1, Deletion as track 2, Insertion as track 3 and SNP as inner track 4 while in other images Substitution as outer track 1 and SNP as inner track 2, (c) SNPs present in more than 10% of SARS-CoV-2 population for Global and India, (d) Screenshots of the web application before and after executing search query
*****
NB/KGS/(DST Media Cell)

